Result browser
This panel is for visualizing the information collected and/or
predicted for the submitted protein.
-
Summary tab
Basic informations about the entry: such as gene name (if available), cross references, the reliability of the predicted topology, the number of transmembrane segments, and the download button for the resulting xml file. -
XML
The raw xml description of the given entry. The XSD schema definition of the xml files can be found here. -
1D tab
Color-coded amino acid sequence of the given human transmembrane protein based on the determined or predicted topology (red: inside/cytosolic; yellow: membrane; orange: re-entrant loop; blue: outside/extra-cytosolic region). -
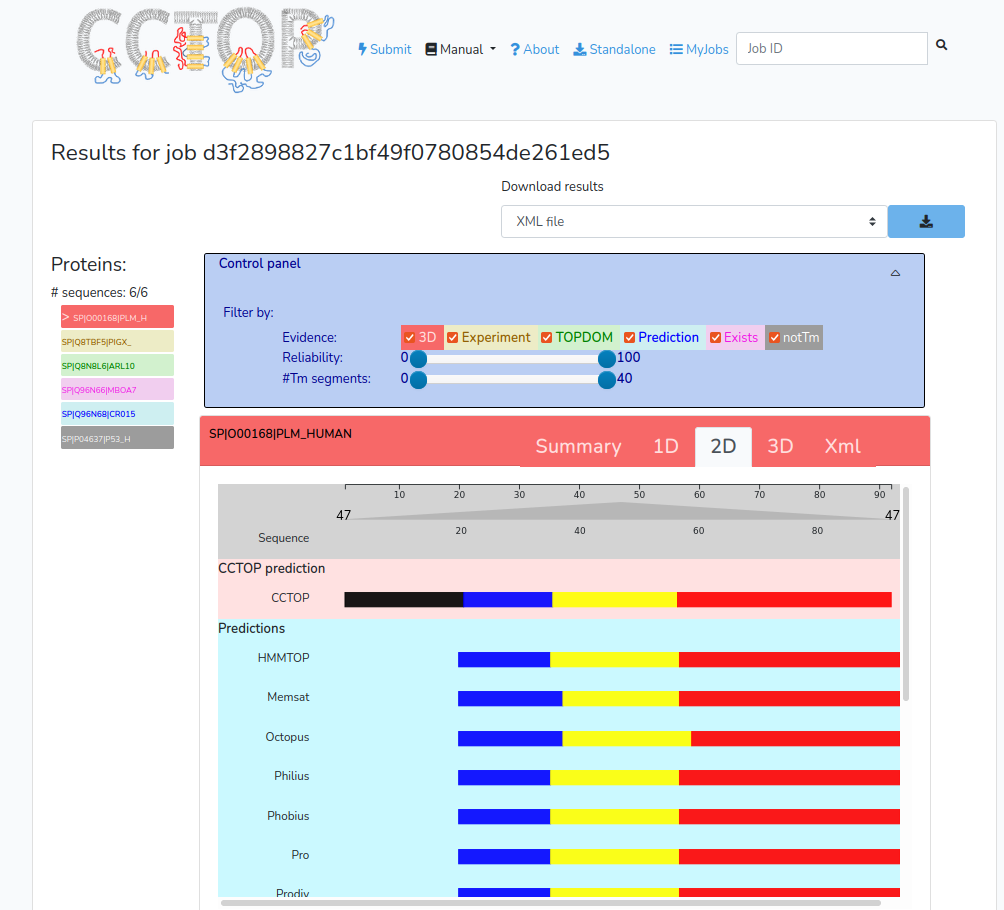
2D tab
A graphical representation of the determined and predicted topology of the given entry. The graph consists of three parts: the final CCTOP prediction, the results of the various topology and topography prediction methods and the collected constraints aligned to the amino acid sequence of the given entry.
The x-axis on the graph is the position in the given sequence, lower red lines represent inside/cytosolic regions, yellow rectangles code transmembrane regions, orange rectangles show membrane re-entrant loops and upper blue lines represent outside/extra-cytosolic regions. Thin gray lines indicate regions, where the topology information is not predicted/determined. -
3D tab
This tab is selectable for entries of which have solved 3D structure. It contains the 3D representation of the structure of the given entry positioned in the membrane bilayer. For the 3D representation, the JSMol is used. The membrane orientation is determined by the TMDET algorithm, data are taken from PDBTM database.
All: #visitors: 2899403, #seqs: 5751655 .:|:.
Last week: 56 .:|:.
Current load: 0